I am a PhD researcher in the Database Architectures group @ CWI. I am working on improving the efficiency of vector indexing and (filtered) search in data systems.
Contributions I am very proud of:
- ALP: an algorithm for floating-point data compression. ALP is used in DuckDB, FastLanes, Vortex, and (soon) Parquet. I also contributed ALP to DuckDB.
- SuperKMeans: an algorithm for super-fast clustering of vector embeddings.
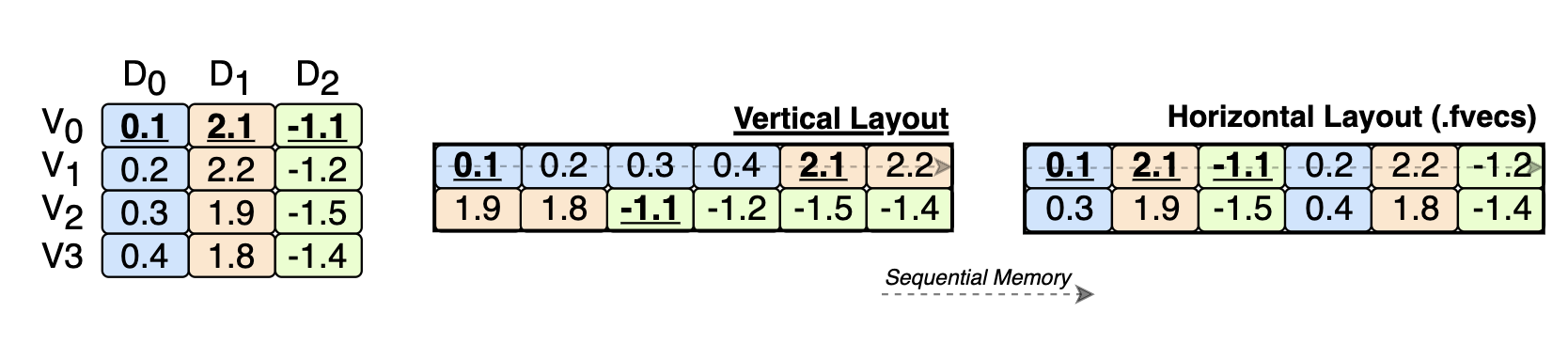
- PDX: a data layout for vector embeddings that speedup vector search.
I am also an online educator: +2M views on YouTube, +12K students on Udemy
News
- [2026/01] Excited to go back to my PhD after interning @ Google IR team!
Contact: [email protected]